Cleaning Data with stringr
Introduction
Occassionally, when we are using a set of data, there may be mistakes. Before we can really make any decisions or visualizations using that data, we must ensure it is as accurate as possible. In this post, we will look at using the R library stringr to filter out unwanted data.
Basic Example
For this example, we will be using a data set from (Kaggle.com)[https://www.kaggle.com/orangutan/exploratory-analysis-of-the-korean-war] that contains all the US deaths from the Korean War. First, we import our libraries and look at the first few rows of data.
library(dplyr)
library(stringr)
#read in the data
deaths<-read.csv("KoreanConflict.csv", header=TRUE, stringsAsFactors=FALSE)
head(deaths)## SERVICE_TYPE SERVICE_CODE ENROLLMENT BRANCH RANK
## 1 V L ACTIVE - GUARD/RESERVE AIR FORCE CAPT
## 2 R K ACTIVE - REGULAR ARMY PVT
## 3 R K ACTIVE - REGULAR ARMY PFC
## 4 V L ACTIVE - GUARD/RESERVE ARMY 2LT
## 5 R K ACTIVE - REGULAR ARMY CPL
## 6 R K ACTIVE - REGULAR ARMY PFC
## PAY_GRADE POSITION BIRTH_YEAR SEX HOME_CITY
## 1 O03 1917 M NEW YORK
## 2 E02 FOOD SERVICE APPRENTICE 1927 M UNKNOWN
## 3 E03 HEAVY WEAPONS INFANTRYMAN 1932 M UNKNOWN
## 4 O01 INFANTRY UNIT COMMANDER 1929 M UNKNOWN
## 5 E04 LIGHT WEAPONS INFANTRY LEADER 1932 M UNKNOWN
## 6 E03 LIGHT WEAPONS INFANTRYMAN 1929 M UNKNOWN
## HOME_COUNTY NATIONALITY STATE_CODE HOME_STATE MARITAL_STATUS ETHNICITY
## 1 NEW YORK US NY NEW YORK MARRIED WHITE
## 2 OCONEE US GA GEORGIA UNKNOWN WHITE
## 3 BIBB US GA GEORGIA UNKNOWN WHITE
## 4 COAHOMA US MS MISSISSIPPI UNKNOWN WHITE
## 5 DICKEY US ND NORTH DAKOTA UNKNOWN WHITE
## 6 NELSON US ND NORTH DAKOTA UNKNOWN WHITE
## ETHNICITY_1 ETHNICITY_2 DIVISION INCIDENT_DATE
## 1 NOT SPECIFIED WHITE 93 BOMB SQ 19 BOMB GP 19510412
## 2 NOT SPECIFIED WHITE 29 RGT CMBT TEAM 19500727
## 3 NOT SPECIFIED WHITE 5 RGT 1 CAV DIV 19510316
## 4 NOT SPECIFIED WHITE 32 INF 7 DIV 19530122
## 5 NOT SPECIFIED WHITE 14 INF 25 DIV 19530529
## 6 NOT SPECIFIED WHITE 5 RGT 1 CAV DIV 19510606
## FATALITY_YEAR FATALITY_DATE HOSTILITY_CONDITIONS FATALITY
## 1 1951 20010402 H DECLARED DEAD
## 2 1950 19500727 H KILLED IN ACTION
## 3 1951 19510316 H KILLED IN ACTION
## 4 1953 19530122 H KILLED IN ACTION
## 5 1953 19530529 H KILLED IN ACTION
## 6 1951 19510606 H KILLED IN ACTION
## BURIAL_STATUS
## 1 Y
## 2 Y
## 3 Y
## 4 Y
## 5 Y
## 6 YIt appears that the INCIDENT_DATE field is formatted like so: YYYYMMDD. To check this, let’s compare the total number of rows in the database with how many rows fit that format. To keep things simple, we will use a regular expression to check that the data contains only 8 numbers.
#Get the total number of records int the data
count(deaths) #36,574## # A tibble: 1 x 1
## n
## <int>
## 1 36574#compare INCIDENT_DATE against the regular expression and get the count of records
fullDateRegEx = "^\\d{8}$"
df<-deaths%>%
filter(str_detect(deaths$INCIDENT_DATE, fullDateRegEx)==TRUE)
count(df) #33,370## # A tibble: 1 x 1
## n
## <int>
## 1 33370Looking further into the data, we can see that the FATALITY column also appears to hold a date in the format we want, and only shows that when INCIDENT_DATE is invalid. Using a simple for loop, we can check to see if FATALITY is valid when INCIDENT_DATE is not, and store the date in INCIDENT_DATE
for (i in 1:dim(deaths)) {
#if INCIDENT_DATE is bad, but FATALITY is good, replace it
if (!str_detect(deaths$INCIDENT_DATE[i], fullDateRegEx) & str_detect(deaths$FATALITY[i], fullDateRegEx)) {
#print(paste(i, "There is a mistake here"))
deaths$INCIDENT_DATE[i]<-deaths$FATALITY[i]
}
}## Warning in 1:dim(deaths): numerical expression has 2 elements: only the
## first used#Check out counts again
df<-deaths%>%
filter(str_detect(deaths$INCIDENT_DATE, fullDateRegEx)==TRUE)
count(df) #36,511## # A tibble: 1 x 1
## n
## <int>
## 1 36511This is an acceptable number of valid records (36,511 out of 36,574), so we can just filter out the remaining bad data and begin to use our data for visualizations and decision making.
deaths<-deaths%>%
filter(str_detect(deaths$INCIDENT_DATE, fullDateRegEx)==TRUE)Visualization
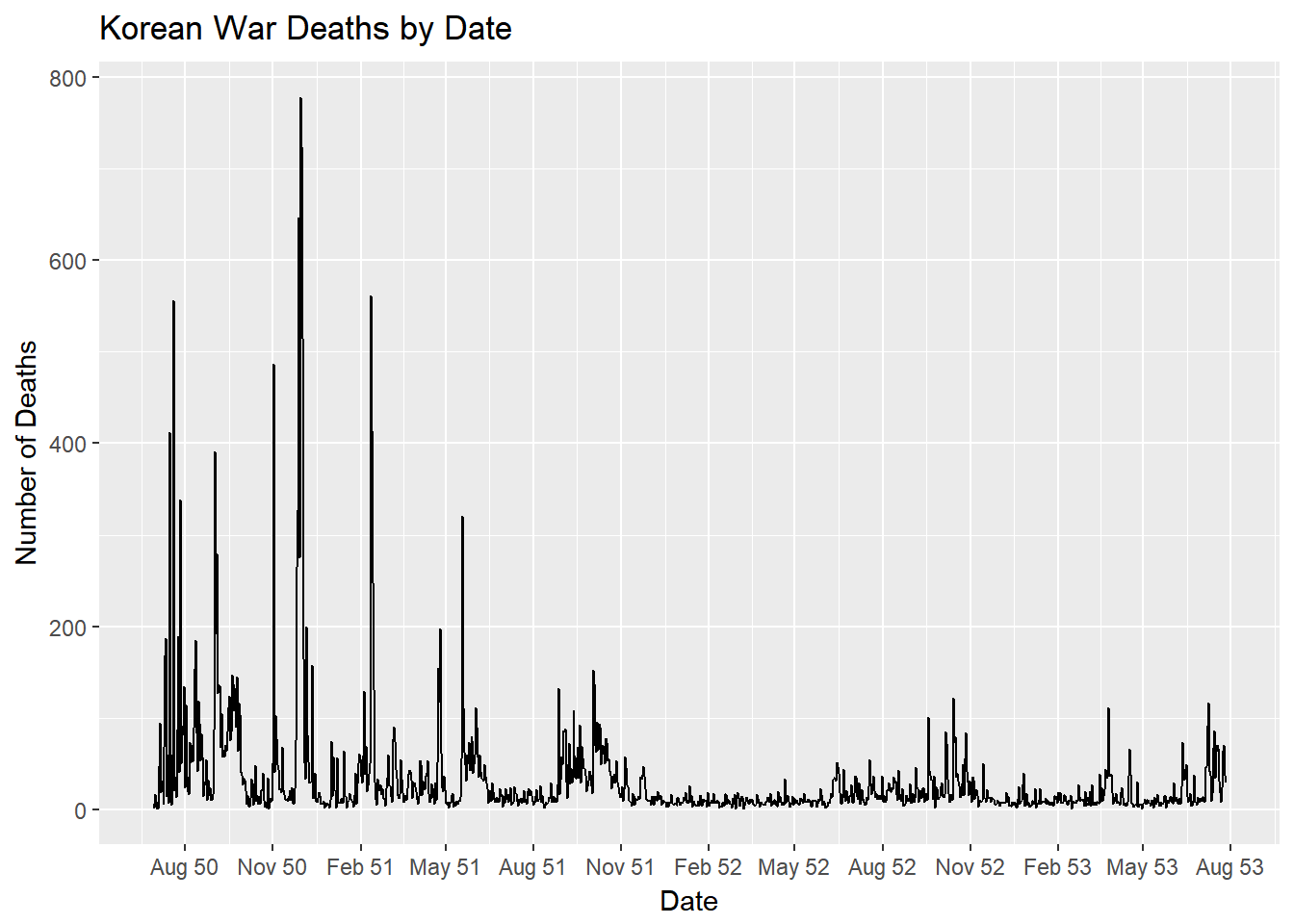
Now we can plot out a simple line chart of deaths by date using this newly cleaned data set.
library(ggplot2)
library(lubridate) #for parsing and dealing with dates
deathsDf<-deaths%>%
group_by(INCIDENT_DATE)%>%
summarize(num_deaths=n())%>%
mutate(date=ymd(INCIDENT_DATE))%>%
filter(date<='1953-07-27')%>%
select(INCIDENT_DATE, date, num_deaths)
#look at number of deaths by date
ggplot()+
geom_line(data=deathsDf, aes(x=date,y=num_deaths))+
ggtitle("Korean War Deaths by Date")+
xlab("Date")+
ylab("Number of Deaths")+
scale_x_date(date_breaks="3 months", date_labels="%b %y")